|
chr14_-_103989195
|
40.647
|
NM_001823
|
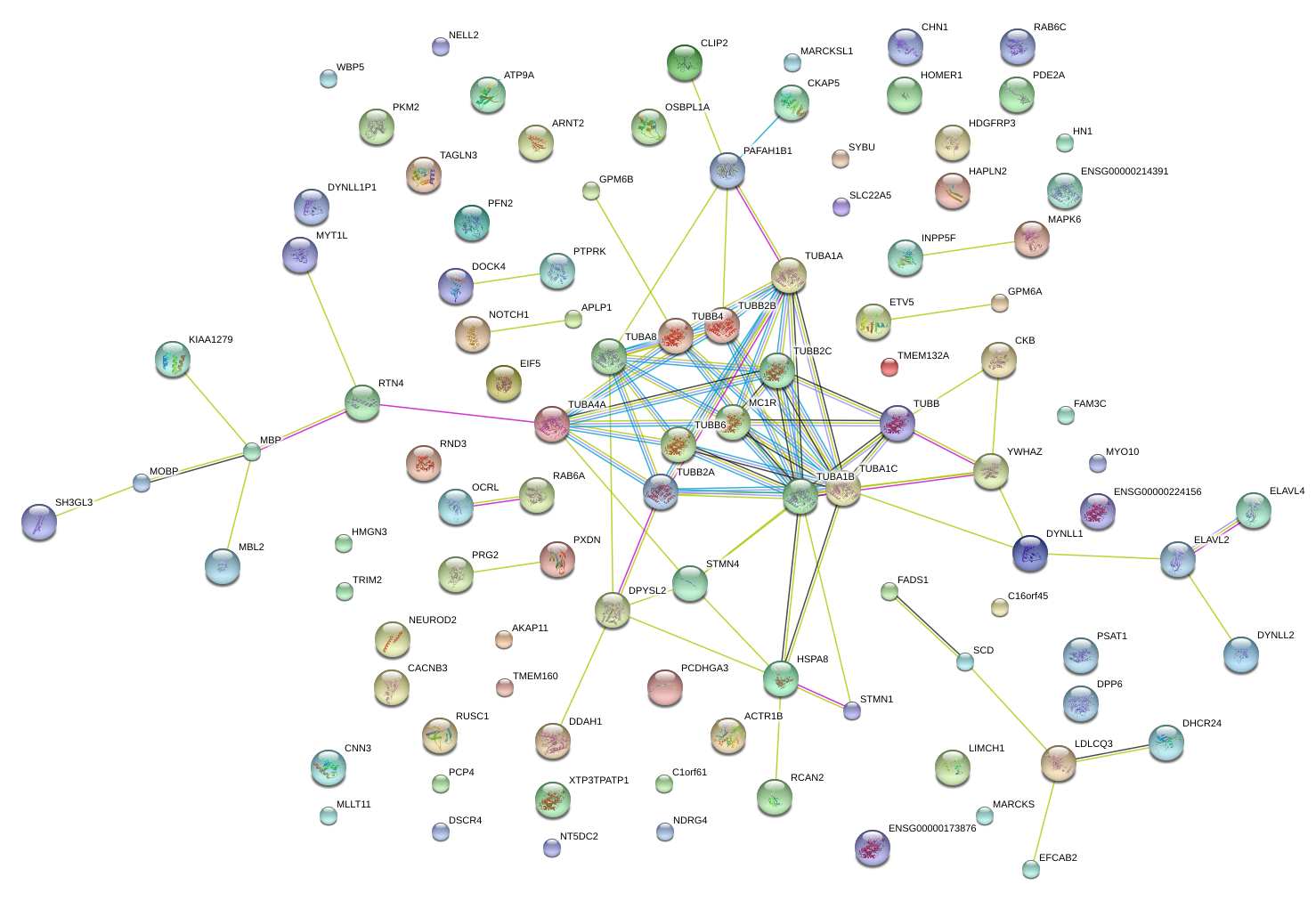
CKB
|
creatine kinase, brain
|
|
chr14_-_103989112
|
40.053
|
|
CKB
|
creatine kinase, brain
|
|
chr6_-_128841432
|
30.126
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr3_-_149688886
|
27.767
|
|
PFN2
|
profilin 2
|
|
chr1_-_26232890
|
27.246
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr18_-_74729049
|
24.832
|
NM_001025081
NM_001025090
NM_001025092
NM_002385
|
MBP
|
myelin basic protein
|
|
chr18_-_74728963
|
24.498
|
|
MBP
|
myelin basic protein
|
|
chr2_-_175869910
|
24.059
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr12_+_120933911
|
23.698
|
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr19_+_36359346
|
21.756
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr1_-_85930610
|
21.245
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr1_-_26232643
|
20.675
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr1_+_151032866
|
19.575
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr8_+_26435339
|
19.039
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr1_+_151032880
|
18.881
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr5_+_140739589
|
18.730
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr15_+_52311408
|
18.709
|
NM_002748
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr12_+_120933832
|
18.314
|
NM_001037495
NM_003746
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr3_+_39509069
|
17.746
|
NM_182935
|
MOBP
|
myelin-associated oligodendrocyte basic protein
|
|
chr15_+_52311438
|
17.697
|
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr2_+_149633173
|
17.265
|
|
|
|
|
chr16_+_89988416
|
16.528
|
NM_001197181
|
TUBB3
|
tubulin, beta 3 class III
|
|
chr15_-_83876769
|
16.517
|
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr5_+_74632992
|
16.511
|
NM_000859
NM_001130996
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase
|
|
chr1_+_50571963
|
16.348
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr11_-_46867840
|
16.291
|
NM_001008938
NM_014756
|
CKAP5
|
cytoskeleton associated protein 5
|
|
chr12_+_120933916
|
16.260
|
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr3_-_149688638
|
16.202
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr9_+_80911990
|
16.048
|
NM_021154
NM_058179
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr15_+_52311432
|
15.819
|
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr17_-_1248346
|
15.487
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr1_+_151032683
|
15.290
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_-_156399183
|
14.699
|
NM_006365
|
C1orf61
|
chromosome 1 open reading frame 61
|
|
chr3_-_185826748
|
14.523
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr6_-_46293404
|
14.374
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr2_-_55277592
|
14.159
|
|
RTN4
|
reticulon 4
|
|
chr15_-_83876165
|
13.923
|
NM_016073
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr4_-_176733901
|
13.742
|
|
GPM6A
|
glycoprotein M6A
|
|
chr5_+_74633043
|
13.707
|
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase
|
|
chr2_-_55277421
|
13.657
|
|
RTN4
|
reticulon 4
|
|
chr10_+_102106771
|
13.519
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr2_-_55277672
|
13.427
|
|
RTN4
|
reticulon 4
|
|
chr12_-_49582296
|
13.352
|
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr2_-_2334887
|
13.294
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chrX_-_13835187
|
13.105
|
|
GPM6B
|
glycoprotein M6B
|
|
chr2_-_55277653
|
13.096
|
|
RTN4
|
reticulon 4
|
|
chr2_-_55277459
|
13.077
|
|
RTN4
|
reticulon 4
|
|
chr2_-_55277620
|
13.035
|
|
RTN4
|
reticulon 4
|
|
chr2_-_55277698
|
12.800
|
NM_020532
NM_153828
NM_207520
|
RTN4
|
reticulon 4
|
|
chr15_+_80696569
|
12.774
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr3_+_111717585
|
12.680
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr1_-_55352893
|
12.656
|
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr17_-_37764165
|
12.491
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr8_-_101965554
|
12.408
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chrX_-_13835313
|
12.290
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr16_+_58534045
|
12.119
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr6_+_114178516
|
11.942
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr8_-_101965601
|
11.938
|
NM_003406
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr1_-_95392449
|
11.925
|
|
CNN3
|
calponin 3, acidic
|
|
chr11_+_60691911
|
11.899
|
NM_017870
NM_178031
|
TMEM132A
|
transmembrane protein 132A
|
|
chr1_-_32801606
|
11.685
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr10_+_70748476
|
11.639
|
NM_015634
|
KIAA1279
|
KIAA1279
|
|
chr6_-_79944397
|
11.632
|
NM_001201362
NM_001201363
NM_004242
NM_138730
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr1_-_95392501
|
11.626
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr16_+_15528324
|
11.414
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr1_-_55352920
|
11.381
|
NM_014762
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr4_+_41614911
|
11.350
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr7_-_121036320
|
11.331
|
|
FAM3C
|
family with sequence similarity 3, member C
|
|
chr8_-_101965096
|
11.313
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr17_-_73150493
|
11.206
|
|
HN1
|
hematological and neurological expressed 1
|
|
chr7_-_111846384
|
11.185
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr1_+_155294217
|
11.136
|
NM_014328
|
RUSC1
|
RUN and SH3 domain containing 1
|
|
chr12_-_49582800
|
10.918
|
NM_006009
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr21_+_41239346
|
10.905
|
NM_006198
|
PCP4
|
Purkinje cell protein 4
|
|
chr8_-_27115935
|
10.888
|
|
STMN4
|
stathmin-like 4
|
|
chr5_-_16936371
|
10.866
|
NM_012334
|
MYO10
|
myosin X
|
|
chrX_+_102611379
|
10.850
|
NM_001006612
NM_001006613
NM_001006614
NM_016303
|
WBP5
|
WW domain binding protein 5
|
|
chr1_-_26233367
|
10.848
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr6_-_79944324
|
10.818
|
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr5_+_140797243
|
10.759
|
NM_018927
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr1_-_55352818
|
10.662
|
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chrX_-_13835012
|
10.536
|
|
GPM6B
|
glycoprotein M6B
|
|
chr5_-_78809658
|
10.384
|
NM_004272
|
HOMER1
|
homer homolog 1 (Drosophila)
|
|
chr1_+_156589085
|
10.313
|
NM_021817
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2
|
|
chr6_-_3227776
|
10.252
|
NM_178012
|
TUBB2B
|
tubulin, beta 2B class IIb
|
|
chr1_+_155293727
|
10.228
|
NM_001105205
|
RUSC1
|
RUN and SH3 domain containing 1
|
|
chr8_-_101965194
|
10.221
|
NM_001135699
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr6_+_30688085
|
10.023
|
|
TUBB
|
tubulin, beta class I
|
|
chrX_-_13835213
|
9.972
|
|
GPM6B
|
glycoprotein M6B
|
|
chr11_-_61584228
|
9.916
|
|
FADS1
|
fatty acid desaturase 1
|
|
chr2_-_151344145
|
9.910
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr5_+_140729723
|
9.885
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr12_+_49212253
|
9.882
|
NM_000725
NM_001206915
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr15_-_72523713
|
9.790
|
NM_001206796
NM_001206797
NM_001206798
NM_002654
NM_182470
NM_182471
|
PKM2
|
pyruvate kinase, muscle
|
|
chr2_-_132121730
|
9.785
|
NM_001077637
|
WTH3DI
|
RAB6C-like
|
|
chr1_-_55352881
|
9.654
|
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr11_-_122932806
|
9.587
|
|
HSPA8
|
heat shock 70kDa protein 8
|
|
chr20_-_50384899
|
9.544
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr11_-_72380107
|
9.513
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr2_+_130737234
|
9.504
|
NM_032144
|
RAB6C
|
RAB6C, member RAS oncogene family
|
|
chr7_+_73703704
|
9.412
|
NM_003388
NM_032421
|
CLIP2
|
CAP-GLY domain containing linker protein 2
|
|
chr8_-_27115897
|
9.251
|
NM_030795
|
STMN4
|
stathmin-like 4
|
|
chr5_+_140797270
|
9.215
|
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr20_-_50384861
|
9.214
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr7_-_121036355
|
9.076
|
NM_001040020
NM_014888
|
FAM3C
|
family with sequence similarity 3, member C
|
|
chr12_-_45269953
|
9.059
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr8_-_110693233
|
9.055
|
NM_001099745
NM_001099746
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr14_+_103800534
|
9.031
|
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr13_+_42846255
|
9.030
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chr12_-_45270157
|
8.998
|
NM_001145108
NM_006159
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr12_-_45270071
|
8.950
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr4_+_154125589
|
8.918
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr10_+_121578768
|
8.836
|
NM_001243194
|
INPP5F
|
inositol polyphosphate-5-phosphatase F
|
|
chr7_+_153584383
|
8.830
|
NM_001039350
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr18_-_21891469
|
8.774
|
NM_001242508
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr3_-_52567809
|
8.767
|
|
NT5DC2
|
5'-nucleotidase domain containing 2
|
|
chr15_+_84115979
|
8.747
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr19_-_47551881
|
8.658
|
NM_017854
|
TMEM160
|
transmembrane protein 160
|
|
chr16_-_19533285
|
8.503
|
NM_016641
|
GDE1
|
glycerophosphodiester phosphodiesterase 1
|
|
chr11_-_122932844
|
8.465
|
NM_006597
NM_153201
|
HSPA8
|
heat shock 70kDa protein 8
|
|
chr19_+_47104389
|
8.395
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chrX_+_21958714
|
8.385
|
NM_004595
|
SMS
|
spermine synthase
|
|
chr11_+_63953630
|
8.293
|
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr7_+_128379428
|
8.258
|
|
CALU
|
calumenin
|
|
chr6_+_123100645
|
8.208
|
NM_001446
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr7_+_128379452
|
8.186
|
|
CALU
|
calumenin
|
|
chr17_-_9940004
|
8.150
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr7_+_128379400
|
8.143
|
|
CALU
|
calumenin
|
|
chr3_-_52567708
|
8.141
|
NM_001134231
|
NT5DC2
|
5'-nucleotidase domain containing 2
|
|
chr5_-_146435589
|
8.100
|
NM_181674
NM_181676
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr4_-_87028805
|
8.093
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr20_+_30193083
|
7.983
|
NM_002165
NM_181353
|
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein
|
|
chr5_-_43313581
|
7.924
|
NM_001098272
NM_002130
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr7_+_128379395
|
7.708
|
|
CALU
|
calumenin
|
|
chr3_+_111718006
|
7.700
|
NM_001008273
|
TAGLN3
|
transgelin 3
|
|
chr18_-_55289014
|
7.627
|
|
NARS
|
asparaginyl-tRNA synthetase
|
|
chr2_+_159313466
|
7.605
|
NM_001005476
NM_003628
|
PKP4
|
plakophilin 4
|
|
chr8_+_109455874
|
7.575
|
|
TTC35
|
tetratricopeptide repeat domain 35
|
|
chr17_-_39684585
|
7.558
|
NM_002276
|
KRT19
|
keratin 19
|
|
chr6_+_121756744
|
7.533
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr18_-_55288951
|
7.483
|
|
NARS
|
asparaginyl-tRNA synthetase
|
|
chr7_+_5632453
|
7.455
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr5_+_140213968
|
7.427
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr4_+_156680125
|
7.398
|
NM_000857
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr17_-_39684499
|
7.376
|
|
KRT19
|
keratin 19
|
|
chr1_+_36621751
|
7.367
|
NM_018067
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr1_-_243418675
|
7.363
|
NM_001042404
NM_001042405
NM_014812
|
CEP170
|
centrosomal protein 170kDa
|
|
chrM_+_4770
|
7.330
|
|
|
|
|
chr7_+_129710336
|
7.330
|
NM_014997
|
KLHDC10
|
kelch domain containing 10
|
|
chr7_-_131241321
|
7.292
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr16_+_8768403
|
7.198
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr19_+_52693229
|
7.188
|
|
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha
|
|
chr11_-_66115016
|
6.995
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr1_-_153643417
|
6.992
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr1_+_200708604
|
6.941
|
NM_203459
|
CAMSAP2
|
calmodulin regulated spectrin-associated protein family, member 2
|
|
chr11_+_63953666
|
6.935
|
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr17_-_1303402
|
6.930
|
|
YWHAE
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide
|
|
chr1_-_197115566
|
6.896
|
NM_001206846
NM_018136
|
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila)
|
|
chr20_+_17207669
|
6.857
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr16_-_74808474
|
6.774
|
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr1_-_94702916
|
6.770
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr1_+_36023389
|
6.730
|
NM_001014839
NM_001014841
NM_014284
|
NCDN
|
neurochondrin
|
|
chr10_-_126107454
|
6.716
|
|
OAT
|
ornithine aminotransferase
|
|
chr1_-_153643474
|
6.714
|
NM_004515
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr7_+_129710401
|
6.698
|
|
KLHDC10
|
kelch domain containing 10
|
|
chr22_-_50523737
|
6.682
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr1_-_153643433
|
6.674
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr4_+_83351725
|
6.665
|
NM_021204
|
ENOPH1
|
enolase-phosphatase 1
|
|
chr7_-_44887697
|
6.642
|
NM_012412
NM_138635
NM_201436
NM_201516
NM_201517
|
H2AFV
|
H2A histone family, member V
|
|
chr16_+_22524843
|
6.641
|
NM_001135865
|
LOC100132247
NPIPL3
|
nuclear pore complex interacting protein related gene
nuclear pore complex interacting protein-like 3
|
|
chr16_-_74808699
|
6.611
|
NM_024306
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr8_+_82192717
|
6.577
|
NM_001444
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated)
|
|
chr2_-_61765345
|
6.577
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr2_-_61765367
|
6.567
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr1_-_246580710
|
6.549
|
NM_022743
|
SMYD3
|
SET and MYND domain containing 3
|
|
chr1_-_153643428
|
6.545
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr11_+_63953586
|
6.505
|
NM_006819
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr18_-_55289026
|
6.488
|
NM_004539
|
NARS
|
asparaginyl-tRNA synthetase
|
|
chr4_-_5890142
|
6.412
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr22_-_50523780
|
6.382
|
NM_139202
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr10_-_123356158
|
6.346
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr16_+_6069094
|
6.330
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr19_-_55919324
|
6.321
|
NM_014501
|
UBE2S
|
ubiquitin-conjugating enzyme E2S
|
|
chr17_+_40610899
|
6.205
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr7_+_149570004
|
6.202
|
NM_001100592
NM_145230
|
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2
|
|
chr22_-_50523853
|
6.195
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr6_+_83072922
|
6.162
|
NM_006670
|
TPBG
|
trophoblast glycoprotein
|
|
chr4_+_41614933
|
6.155
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr7_+_5632435
|
6.132
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr5_-_43313492
|
6.106
|
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr19_+_45394459
|
6.094
|
NM_001128916
NM_001128917
NM_006114
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr6_-_116575170
|
6.091
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr4_-_176733413
|
6.047
|
|
GPM6A
|
glycoprotein M6A
|
|
chr4_-_5894695
|
6.042
|
NM_001014809
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr11_+_57479994
|
6.019
|
NM_001144012
NM_015959
|
TMX2
|
thioredoxin-related transmembrane protein 2
|
|
chr3_-_52001388
|
6.015
|
NM_020418
NM_001174100
NM_033008
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr19_+_52693334
|
5.986
|
|
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha
|
|
chr4_+_83351765
|
5.977
|
|
ENOPH1
|
enolase-phosphatase 1
|
|
chr1_-_201476248
|
5.977
|
NM_001193570
NM_004078
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chrX_-_77225134
|
5.974
|
NM_001029891
|
PGAM4
|
phosphoglycerate mutase family member 4
|